Research
I. Kinetics of Drug Binding and Mobility in Bilayer Membranes as Revealed by Multinuclear, Dynamic NMR
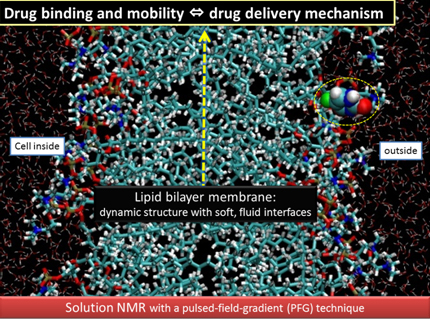
Drug binding to cell membranes is crucial as a primary step of the biological action. Lipid bilayer membranes are dynamic structures where molecules are always fluctuating under physiological conditions. The mechanism of drug deliveries is related to the molecular dynamics in such soft, fluid membrane interface.
The aim of our research is:
To elucidate the mechanism of drug deliveries (DD) in soft, fluid membranes at the molecular level.
Multinuclear, dynamic NMR is mainly used to gain insight into the mechanistic view of DD processes "in a noninvasive manner". Special focus is on how to quantify the drug binding and mobility in membranes in situ.
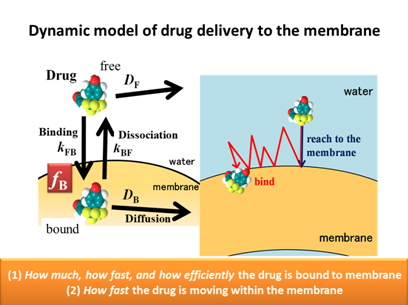
Recently we have succeeded in the simultaneous observation of membrane-bound and free drugs in situ, by applying solution-state NMR in combination with the pulsed-field-gradient (PFG) technique. This enables us to quantify i) in what amount the drug is bound in situ and ii) how fast the drug is moving within the model cell membrane, in relation to the thermal fluctuation of the soft, fluid environment. (see Ref. 1)
The kinetics of membrane binding and dissociation of drugs is also quantified by PFG or 1D NMR, using the solution of Bloch equation with exchange terms. (see Refs. 2-4)
References
- E. Okamura and N. Yoshii, J. Chem. Phys. 129, 215102 (2008).
- N. Yoshii and E. Okamura,Chem.Phys. Lett. 474, 357-361 (2009).
- N. Yoshii and E. Okamura,J. Phys. Chem. B 115, 11074-11080 (2011).
- N. Yoshii, T. Emoto, and E. Okamura, BIOPHYSICS, 7, 105-111(2011).
II. Real-Time In-Cell NMR Spectroscopy of Non-Endocytic Membrane Transport
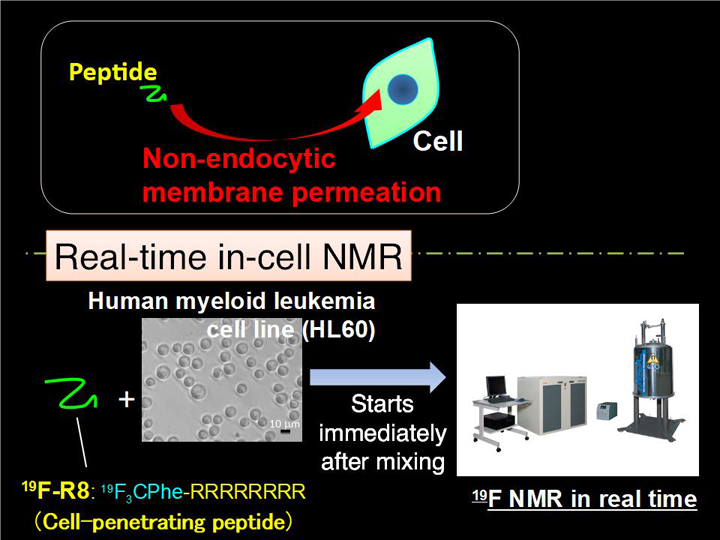
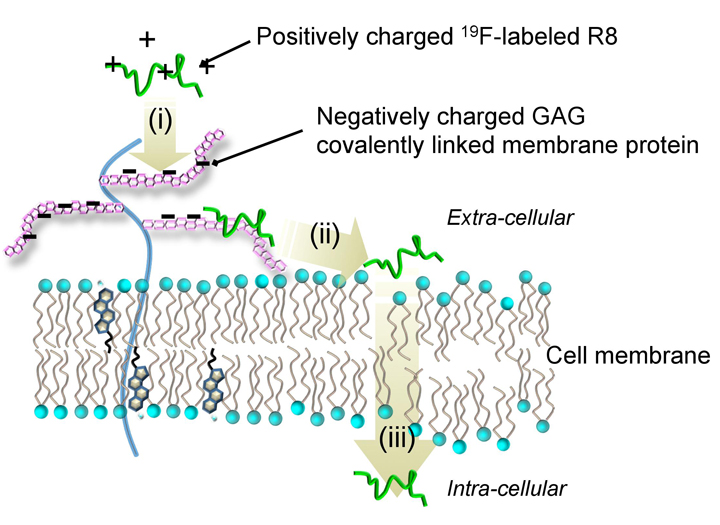
In-situ comprehensive observation and the quantitative analysis of drug deliveries are of significance, but still challenging. We have applied real-time in-cell NMR spectroscopy to investigate the non-endocytic transport of membrane-permeable octaarginine (R8) to living cells (Fig. 2-1). We observed the spontaneous membrane translocation of R8 into a human myeloid leukemia cell line (HL60) with a time resolution in the order of minutes, by introducing 4-trifluoromethyl-L-phenylalanine to R8. 19F NMR successfully detected the real-time R8 transport: (i) the binding to anionic glycosaminoglycans (GAG) at the cell surface, (ii) followed by the penetration into hydrophobic membrane by charge neutralization, and (iii) entry into the cytosol across the membrane (Fig. 2-2). NMR concentration analysis enabled quantification of how much of R8 was staying in the respective translocation processes with time in situ. Taken together, our in-cell NMR results provide the physicochemical rationale for how cationic cell-penetrating peptides traverse hydrophobic membranes.
Reference
Y. Takechi-Haraya, K. Aki, Y. Tohyama, Y. Harano, T. Kawakami, H. Saito, and E. Okamura, Pharmaceuticals, 10, 42 (2017).
III. Lipid Membrane Dynamics in Cell Sized Giant Vesicle by NMR
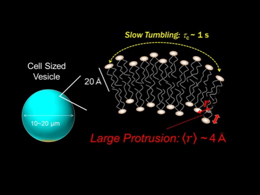
Lipid membrane dynamics in cell sized vesicles (CSVs) in water is characterized by the solution-state NMR. We have successfully prepared CSVs by natural swelling method using osmotic effect. CSVs, larger than 10 μm in diameter, are not only the most suitable model for cell membranes, but also provide great advantages in quantifying the inter-nuclear distance <r> of the lipids by using solution NMR measurement. The slow tumbling motion of the lipid is shown in CSVs of 10-20 μm-diameters, with the rotational correlation time τc of sub-second to second order. The 1H-1H NOE enhancement demonstrates that the distance <r> between lipid headgroup terminal and hydrophobic alkyl chains is estimated to be ~0.4 nm in the fluid phase. The proximity between hydrophilic headgroup and alkyl chains of the lipid is due to the large protrusion in the vertical direction of the CSV surface, despite the little curvature of the membrane. The present study shows the potential of the solution NMR to elucidate molecular dynamics relating to the mechanism of biological functions via the cell membrane.
Reference
Y. Takechi, H. Saito, and E. Okamura, Chem. Phys. Lett., 570, 136-140 (2013).
IV. Spontaneous, Non-Enzymatic Peptide Bond Cleavage of Amino Acid Isomers by Real-Time Solution NMR
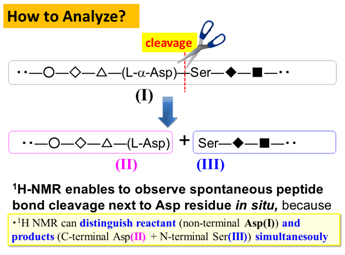
Although L-amino acids were selected as main constituents of peptides and proteins during chemical evolution, D-aspartyl (Asp) residue is found in a variety of living tissues. In particular, D-β-Asp is thought to be stable than any other Asp isomers, and this could be a reason for gradual accumulation in abnormal proteins and peptides to modify their structures and functions.
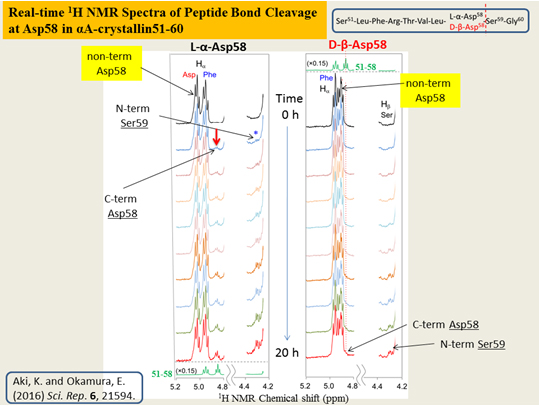
We have compared the spontaneous peptide bond cleavage between Asp isomers, by applying real-time solution-state NMR to eye lens αΑ-crystallin 51―60 fragment, S51LFRTVLD58SG60 and αΒ-crystallin 61―67 analog, F61D62TGLSG67 consisting of L-α- and D-β-Asp 58 and 62, respectively. Kinetic analysis showed how tough the uncommon D-β-Asp residue was against the peptide bond cleavage as compared to natural L-α-Asp. Differences in pKa and conformation between L-α- and D-β-Asp side chains were plausible factors to determine reactivity of Asp isomers. The study, for the first time, provides a rationale to explain less reactivity of D-β-Asp to allow abnormal accumulation.
Reference
K.Aki and E. Okamura, Scientific Reports, 6, 21594 (2016).
